Key Concepts for Data Contributors
If you are submitting data to FaceBase, we recommend that you begin by reviewing the following key concepts and relationships. The diagram here is a high-level representation of the database model (i.e., the way information are structured) in FaceBase.
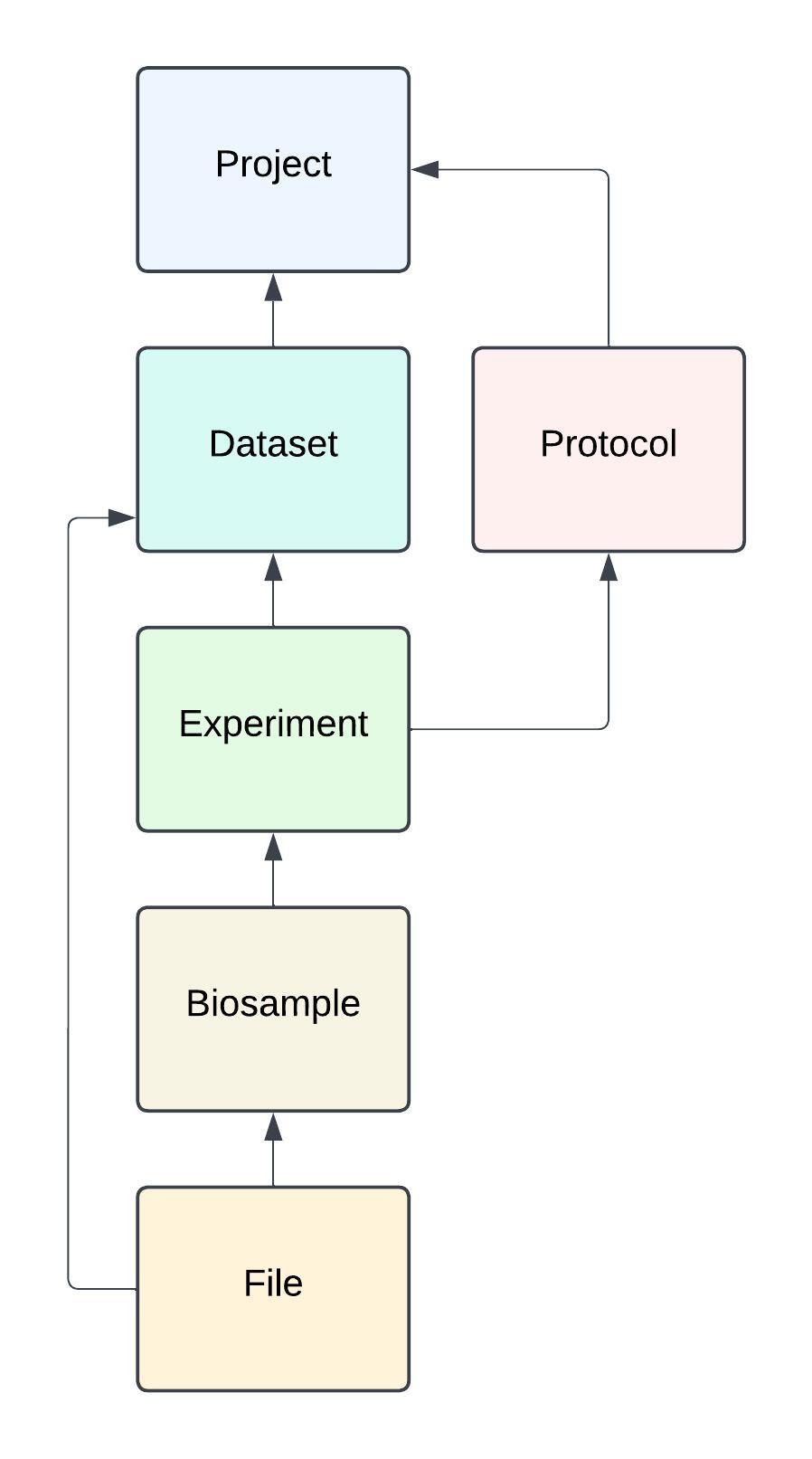
-
Project: represents a research project (e.g., a R01 investigation, etc.). Projects include a set of project investigators and project members (not depicted).
-
Dataset: represents a unit of data collected and submitted to the FaceBase site. Typically, a dataset represents a whole or a logical unit of an investigation (i.e., a study within an overall investigation/project).
-
Protocol: a means of describing the methods used for an experiment. It should include sufficient details to reproduce the results of the experiment. The details may be entered in a rich text editor (online), attached as a document file, or a URL may be given to reference an already published protocol description. We recommend following the Nature Protocol Exchange or similar guidelines for documenting protocols.
-
Experiment (a.k.a., assay): represents an experiment at a fine-grained unit of detail. It is intended to broadly cover multiple “bioinformatics” (i.e., sequencing or array) and imaging (i.e., various forms of microscopy) assay types. An experiment will generally be conducted on multiple biological replicates. An experiment may reference another experiment as its “control.” Additional concepts (not depicted) for specialized experiment types include Enhancer for enhancer reporter assays with transgenic information and Clinical for additional patient details relevant to clinical studies.
-
Replicate: used to create collections of mutant or control biosamples and associate them with an experiment. To keep things simple for data entry and display, the ‘replicate’ has simple numeric columns to record the “biological replicate number” and “technical replicate number.”
-
Biosample: represents the biosample used in an study. There may be many biosamples in a dataset and biosamples are generally grouped together and associated with an Experiment as a collection of biological replicates. Note that FaceBase does not collect physical tissue samples. The ‘biosample’ here is only the metadata used to describe the physical samples used in your experiments.
Vocabulary
FaceBase has adopted external, standardized “vocabulary” a.k.a. “ontology” for most terminology used to describe data. These include experiment types, species, age and development stages, anatomical terms, phenotypes, syndromes, gene nomenclature, and others. Most vocabulary terms are managed by the FaceBase Hub. If you cannot find a term that you need in order to describe your data, please contact us. For additional information, see the NCBO BioPortal.
Currently, we use the following vocabulary:
| Category | Source(s) | Available on FaceBase |
|---|---|---|
| Experiment Type | OBI | View online |
| Species | NCBI Taxonomy | View online |
| Gene | NCBI Gene | View online |
| Phenotype | MP, HPO, OCDM | View online |
| Stage | Varies by species | View online |
| Anatomy | UBERON, OCDM | View online |
| Strain | MGI | View online |
When a needed term is not available in the standard vocabulary, we will work with data submitters to create a new term on FaceBase and promote it to the appropriate upstream vocabulary maintainers for future standardization.
Experiment Types
FaceBase data should be the product of or useful for craniofacial research. See the experiment type terminology for a list of anticipated experiment types for data submissions. Again, if you do not see your experiment type on the list, please contact us so that we can discuss the right terminology to add to the Hub.
Data Types and Formats
Most experiments will produce one or more of the following types of data. We list here the types and formats of data that the Hub accepts. The Hub is always open to discussing the inclusion of additional data types and formats, if they can be of value to craniofacial and dental research. We generally favor data formats that are either “open” (either as a standard or de facto) and for which free or widely used tools are available for using the data files.
- Sequencing Data: “raw” sequencing data (fastq files). These must be gzipped and use the ‘.fastq.gz’ file extension. If you use the common naming scheme to indicate the sequence read number, ‘example_1.fastq.gz’ or ‘example_R2.fastq.gz’, the system will automatically extract the read number from the file name.
- Processed Data: data that are derived from sequencing data through a particular pipeline. Usually fastqc reports (.fastqc.tgz or .fastqc.zip), count files (.count, .tpm, .fpkm), measures in tab-separated format (.tsv), and of course alignment mapping files (.bam) and indexes (.bam.bai).
- Track Data: data that are derived from sequencing or processed data and used in genome browsers, such as BED (.bed), bigBed (.bb), and bigWig (.bw) files.
- Array Data: “raw” microarray data (CEL files). These must be gzipped and use the ‘.CEL.gz’ file extension.
- Imaging Data: high-resolution 3D or 2D imaging data, such as micro-CT accepted in NIfTI format gzipped (.nii.gz), confocal or other microscopy sources in TIFF or OME-TIFF (.tiff or .ome.tiff), and other sources in JPEG (.jpg or .jpeg). Other formats may be considered on an as needed basis.
- Surface Model / Mesh Data: 3D surface models (a.k.a., “polygon mesh” files) that are generally derived from hard tissue imaging data. Currently, we only accept Wavefront OBJ format and it must be gzipped (.obj.gz). If you want these files to be visible online in the Hub’s online mesh viewer, we strongly advise that you limit the size of any model (which may include more than one obj.gz mesh object) to under 10 MB. The entire model must be downloaded to the user’s browser in order to view, and therefore larger models can be prohibitively time consuming to download for users on slower networks. Multiple mesh data entities can be associated with each imaging data entity. Note that human subjects “mesh” files are handled through a completely separate and secure data upload process and released only under approval of our Data Access Committee (DAC).
- Thumbnails: smaller images that give a representation of imaging data. These thumbnails are displayed online in the Hub’s data browser and do not require login for users to view and therefore a good way of showing users enough about the images to encourage them to login (or signup) and download your data. Multiple thumbnails can be associated with each imaging data entity.
Supplementary Data
Finally, the Hub allows for a “supplementary data” category for each dataset. However, this is only to be used for data files that fall outside of the above categories. Please do not submit any of the above data types in this general “file” category. For example, a good use for the generic file category is a spreadsheet that gives additional measures about your imaging or sequencing experiment that are relevant across your entire dataset and are not specific to any one experiment or replicate or file.
Thumbnail Images
Thumbnails can also be uploaded and displayed directly on the main page of your dataset. In order for a thumbnail to appear on the main page of your dataset, you need to reference your dataset from the thumbnail record’s “dataset” field and you need to set the “Show On Dataset” field to “True.”
References to External Data and Visualization
For some types of Datasets, such as Genome Wide Association Studies (GWAS), it may be appropriate to include references to external sources of data and visualization. For example, a GWAS study may have produced or reused genomic data submitted to dbGaP, in addition derived results would have been generated from the original data, and the derived data may have been submitted to LocusZoom for online visualization. In cases such as these, you should enter a Dataset with a clear Description of the data and a thorough Study Design. You should provide accession numbers to data at external repositories (e.g., dbGaP) and Web references to the visualizations of your data using the [label](url) syntax available in the Description field; e.g., [View results for ABC](https://example.org/to/abc). By following these guidelines, users of your data will be able to find your datasets on FaceBase and then navigate to the visualization interface for your data.
Next step, Data Submission Process.