Key Concepts for FaceBase Users
Searching on FaceBase: Faceted Navigation
Our approach to search is more like the experience of shopping online than it is performing a web search.
For example, if you’re shopping for a car online, web search might help narrow down which online resource to use for finding your car - but it will probably not help you find the specific car you want to buy.
The reason is that sites like Google are good for searching for primarily text-based objects like documents and web sites.
But to identify a particular item - like a car - there are specific attributes that you can use to locate something of interest: a specific make or model, cost, at a certain location, etc. These attributes or “facets” classify this object.
This is why in many online shopping sites, you’ll find a filtering sidebar that allows you to pick and choose whichever attributes are interesting to you. You can also change your choices to view different results. This approach is called “faceted navigation”.
This method guides your search through structured data by their classification or characteristics.
Biomedical research involves experiments on a group of model organisms (mice, zebrafish, etc) with particular age stages, there’s at least one anatomical region of interest, and there are different assays (RNA-Seq, microCT, etc). These are examples of the different categories or “facets” of the data.
FaceBase provides guided search based on these facets.
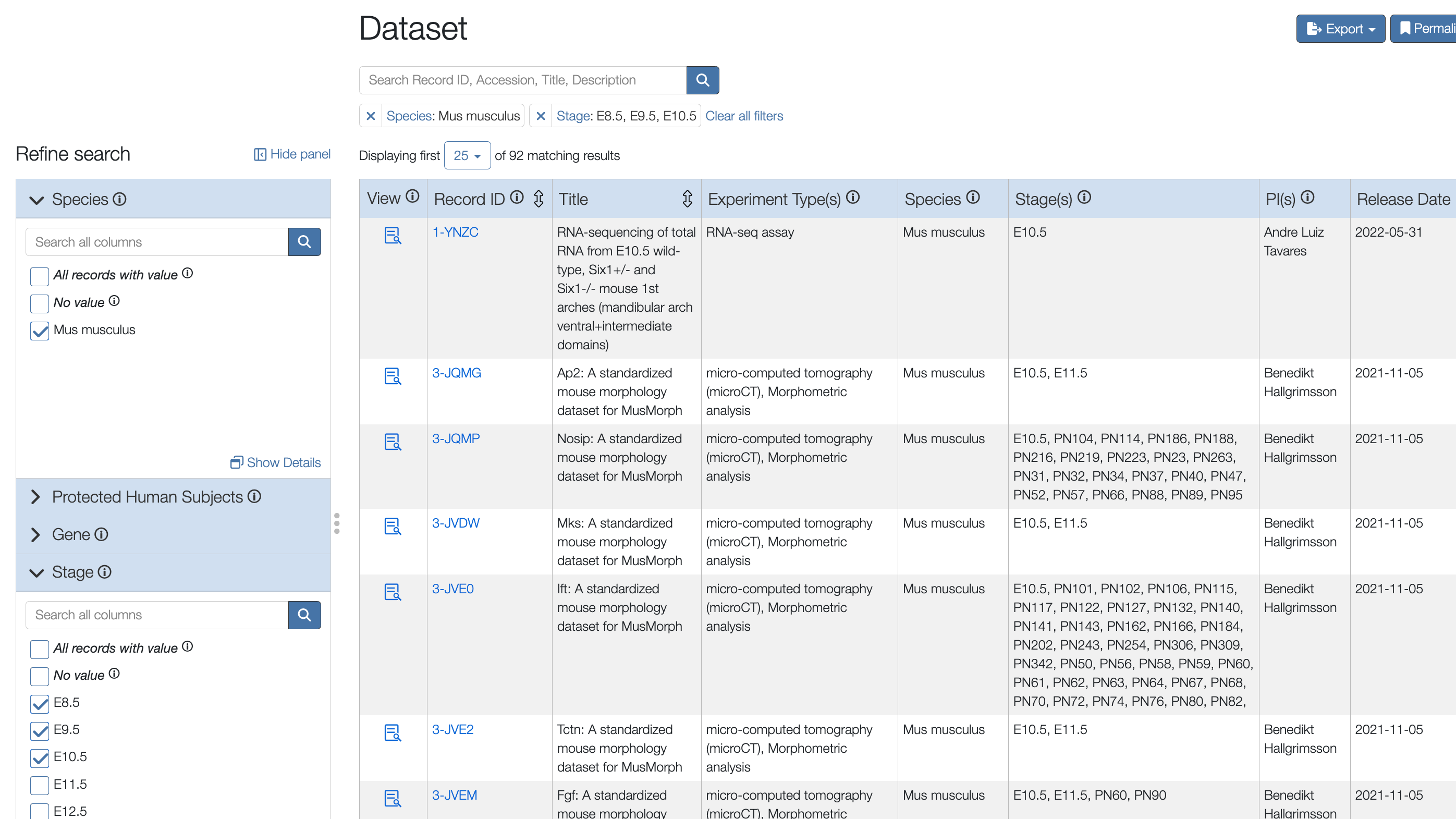
When you perform a search in FaceBase, you’ll see the faceting navigation sidebar on the left, with the main search results on the right. As you explore the available facets and make choices, the main search results update to show what’s available with those combination of facets.
How data is structured in FaceBase (ie. the data model)
Instead of just providing a title, abstract and files, FaceBase data is organized into a hierarchy based on the concepts used in research studies: Projects, Datasets, Experiments and Biosamples:
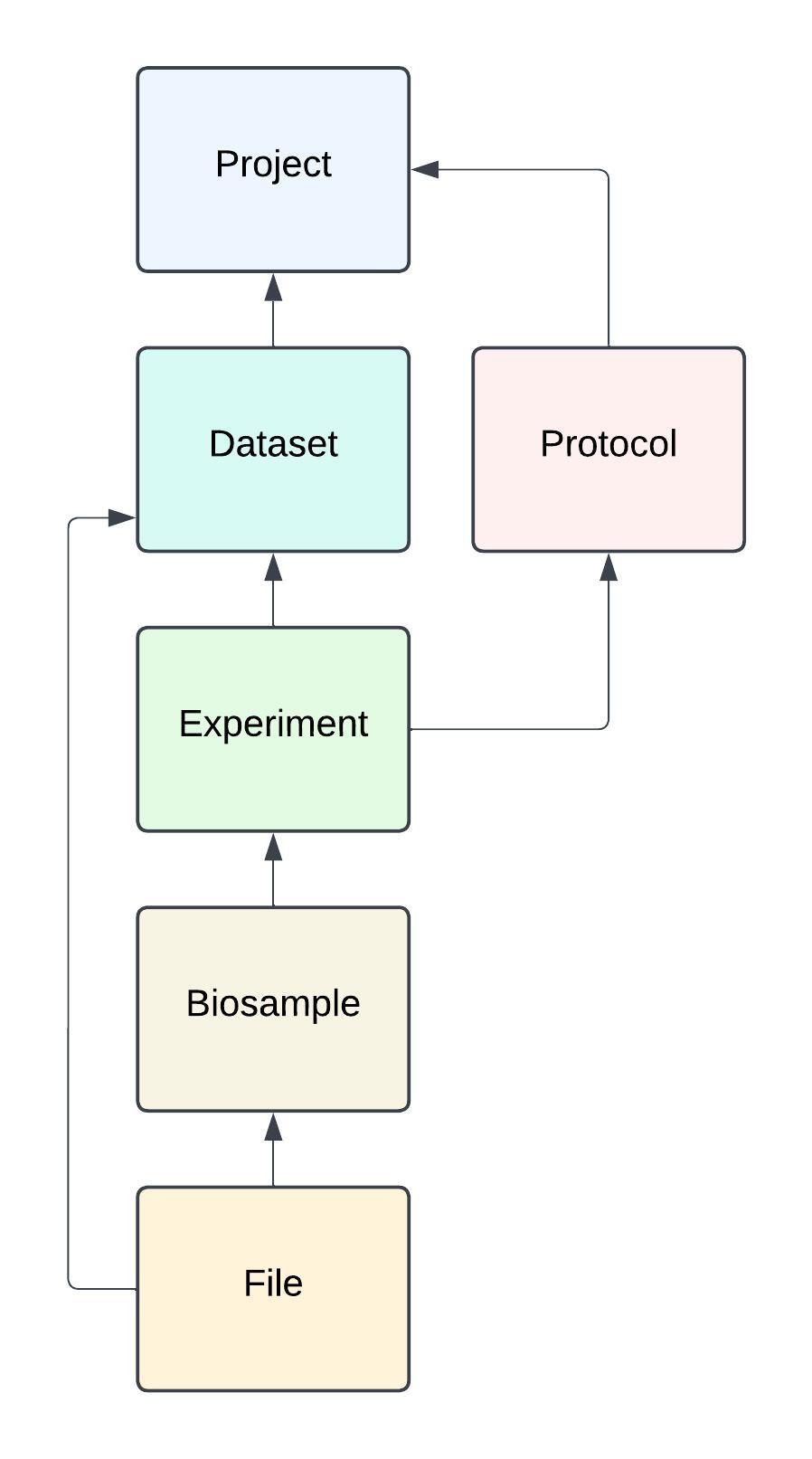
- Projects record the details of the projects that have contributed data in order to provide proper attribution. A Project is equivalent to an R01 investigation for example.
- The resulting data from a project is organized into Datasets which describe a study as a whole.
- Within each dataset is one or more Experiments which are described by their Protocols.
- Each Experiment has Biosamples that describe the biological characteristics of the specimens.
- Files are associated with the specific biosample they were produced from. Which means you know the exact biological specimen where every data file came from, the biological characteristics of the image, raw sequencing read, or other data you’re looking at and the related Experiment.
FAIR Principles
One of the priorities of FaceBase is to provide data that is based on FAIR principles: which makes sure data is findable, accessible, interoperable, and reusable. We enable these through the following practices:
- Faceted navigation allows discovery across disparate data types from varied sources.
- FaceBase emphasizes well-defined metadata and data models, the use of controlled vocabularies and ontologies, and any other ways to include enough measurable metadata for all data entered to allow the appropriate data to show up in searches, and allow others to reproduce and verify experiments.
- We ask data contributors to provide raw, source data in open source formats as our first priority. When that is not possible and the data is desirable, we allow proprietary formats if they are usable using open-source and easily available tools.
- All of our RNA-Seq and ChIP-Seq data are run through a bioinformatics pipeline that standardizes the output for valid cross reference between datasets from different labs.
RIDs
Every data record throughout FaceBase data has its own Resource IDentifier (RID). This is a permanent, citable, globally unique identifier displayed for every data record, much like an Accession Number. For more information on citing with RIDs, go to Sharing and citing data.